Notice: This Wiki is now read only and edits are no longer possible. Please see: https://gitlab.eclipse.org/eclipsefdn/helpdesk/-/wikis/Wiki-shutdown-plan for the plan.
2019 novel Corona Virus Model
This page is under construction
The 2019 novel Corona Virus Model is an all in one archive available on the download page that allows you to install and run a global model to study 2019-nCoV.
Please Observe: The correct epidemiological parameters are not known at this time.
Documentation you should read to install and run the model
Please see the following wiki pages for help
- Importing and Exporting Projects
- using the The STEM UI and Map View
- setting up Simulation Data Logging
- Triggering interventions
Archive and it's contents
Please visit the downloads page and download the single archive. Extract the archive to your local file system. It contains four primary components.
All four of these components must be imported directly into your STEM work space (not nested in another folder). If you already have the GlobalGeography or the SuperContinentExamples projects installed you need not reinstall them (but they are required to run the scenario).
- The project ncov.eclipse.org.coronavirus is the model builder project containing the disease model.
- Project folder CoronaVirus contains the global scenario.
Setting up the model
Once you have installed all four projects and can see them all, un-nested, in the project explorer (in your workspace),
- navigate to the ncov.eclipse.org.coronavirus
- open the models folder.
- make sure you are in the designer perspective
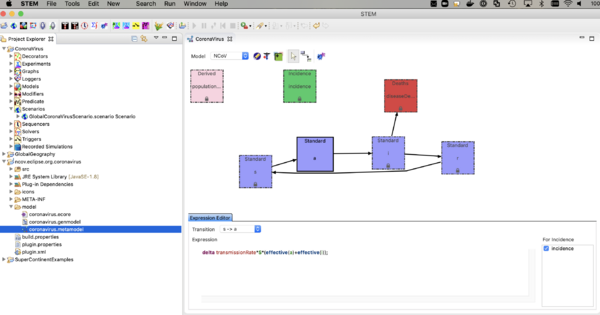
- double click on coronavirus.metamodel
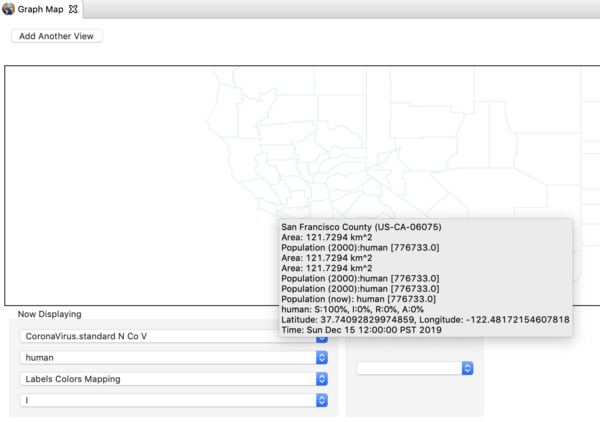
you should the see the compartment model itself as shown below.
The nCoV model is an SAIR model where A represents individuals who are shedding virus (infectious) but asymptomatic. The I compartment represents individuals who are infectious and symptomatic. The incidence (or transition from S -> A, depends on the sum of the fraction infectious in both A and I. Click on the transition and see the expression in the expression editor. You can watch the youtube video on the model builder to see how this works.
The model looks a lot like and SEIR model but there is not incubation state or incubation rate (you could add that if you want to). The reason to explicitly model the A compartment is that you can add interventions (predicates, modifiers, and triggers) that have effect on the I state but not A. The rate individuals move from A -> I is called the "Symptom Appearance Rate" in this model. Next,
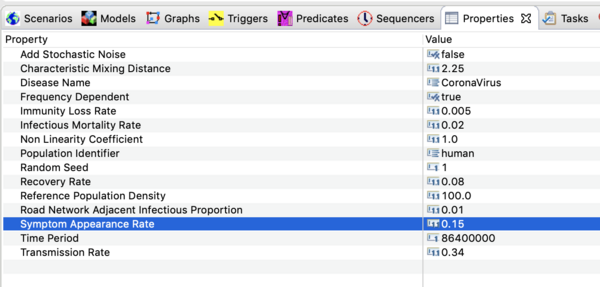
- Navigate to the CoronaVirus project, open the decorators folder, and double click on the disease model (not the infector) to open it in the resource window. Click on it in that window and open the property view below. You should see all the parameter values defined in this model as shown in
the image below.